Given the enormous implications in regulation of cellular function by the modulation of gene expression that regulate gene expression. Transcription regulatory machinenary in Folinic acid calcium salt pentahydrate eukaryotes involves specific transcription factors and transcription inhibitors; proteins that are required to turn on and turn off the expression of particular genes. These considerations point to a possible mechanism of how oxalate differentially regulates the gene expression of such a large number of genes. Given that oxalate is a metabolic end product in humans that cannot be further metabolized, such large scale changes in gene expression in renal epithelial cells in response to high oxalate levels points to an indirect mechanism of action, which may involve the interaction of oxalate with the cell membrane or in intracellular components. The primary site of oxalate action in cell remains unknown. Irrespective of primary site of action, one of the most common means by which cells sense changes is by activating the signal transduction pathways, especially the stress signal pathways. The stress associated signals are transduced through a series of proteins that are activated by phosphorylation/dephosphorylation steps and are finally turned into transcription factors, causing changes in gene expression. Though the present study design does not allow for the identification of activity changes due to phosphorylation, we identified changes in the gene expressions of upstream activators of several signaling pathways. Proteins like Ras, Fas and MKK are highly up-regulated as a result of oxalate exposure. These proteins are known to play important roles in JNK/SAPK signaling and p38 MAPK signaling. These results are in agreement with previous studies, by us and others, that identified an active role for Stress Activated Protein Kinases in oxalate renal cell interactions. We also identified changes in the expression of genes associated with retinoic Acid Receptor Signaling Pathway. Clearly additional studies are required to evaluate the functional consequence of these gene expression changes. In summary, our study is the first attempt at profiling the Genome-wide expression changes in human renal 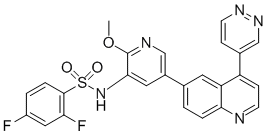 epithelial cells as a result of exposure to oxalate. Results from our study point to complex and intricate mechanisms, including differential gene expression, in renal epithelial cells in response to oxalate exposure. Clearly further studies are required to completely understand the implications of the plethora of changes in gene expression occurring as a result of oxalate exposure in renal epithelial cells. We must separate and characterize the genes that are derived from the by-stander effect and identify the genes whose altered expression is responsible for oxalate nephrotoxicity. Malaria due to Plasmodium falciparum remains a major global health burden and a leading cause of death worldwide among children under five. Benzethonium Chloride Increasing drug resistance, including emerging resistance to the artemisinin drugs, and the declining efficacy of vector control interventions in some populations make the development of effective malaria vaccines an urgent priority. During blood-stage infection, P. falciparum merozoites invade erythrocytes, mediated by the release of invasion ligands from apical organelles that interact with receptors on the erythrocyte surface. The repertoire of invasion ligands includes two major families, the P. falciparum reticulocyte-binding homologues, and erythrocyte binding antigens. The ability of P. falciparum to vary the expression and/or use of EBA and PfRh proteins enables the use of alternate invasion pathways, facilitating immune evasion that enables P. falciparum to cause repeated and chronic infections. Invasion pathways can be broadly classified into two main pathways, sialic acid dependent invasion and SA-independent invasion.
epithelial cells as a result of exposure to oxalate. Results from our study point to complex and intricate mechanisms, including differential gene expression, in renal epithelial cells in response to oxalate exposure. Clearly further studies are required to completely understand the implications of the plethora of changes in gene expression occurring as a result of oxalate exposure in renal epithelial cells. We must separate and characterize the genes that are derived from the by-stander effect and identify the genes whose altered expression is responsible for oxalate nephrotoxicity. Malaria due to Plasmodium falciparum remains a major global health burden and a leading cause of death worldwide among children under five. Benzethonium Chloride Increasing drug resistance, including emerging resistance to the artemisinin drugs, and the declining efficacy of vector control interventions in some populations make the development of effective malaria vaccines an urgent priority. During blood-stage infection, P. falciparum merozoites invade erythrocytes, mediated by the release of invasion ligands from apical organelles that interact with receptors on the erythrocyte surface. The repertoire of invasion ligands includes two major families, the P. falciparum reticulocyte-binding homologues, and erythrocyte binding antigens. The ability of P. falciparum to vary the expression and/or use of EBA and PfRh proteins enables the use of alternate invasion pathways, facilitating immune evasion that enables P. falciparum to cause repeated and chronic infections. Invasion pathways can be broadly classified into two main pathways, sialic acid dependent invasion and SA-independent invasion.
Author Archives: Metabolism
The viability of umbilical cord as a stem cell source is supported by the reports of several studies
Trp155 is not conserved in any if the bacterial genomes available at present, while the appearance of Phe at equivalent position to Phe150 is rare. Thermophilic enzymes frequently present increased number of clustered aromatic residues, which are often mutated to Leu in the mesophilic counterparts, as is the case in cvPAH. Surfaceexposed small aromatic clusters, often located close to the active sites, have been found to confer an entropic advantage over mesophilic analogues through generation of low-frequency motions. Based on the high sequence identity with the other PAH enzymes of known 3D-structure, Trp155 would be located at the start of the helix leading to the iron-coordinating residue Glu167 and might form a stabilizing aromatic cluster with Phe150 and/or Phe92. Furthermore, it is also well established that oligomerization is a strong stabilization mechanism and a large proportion of hyperthermophilic proteins have a higher oligomerization state than their mesophilic counterparts. In this context it is interesting that thermostable lpPAH appears to be dimeric, while other purified and characterized bacterial PAHs are monomeric. The clinical Butenafine hydrochloride symptoms and signs of ARF manifest as a rapid loss of the ability of the kidneys to excrete wastes, concentrate urine, and maintain fluid and electrolyte homeostasis. In pathophysiology, ARF may result from prolonged renal hypoperfusion and renal ischemia or nephrotoxic substances, and is associated with tubular cell death and shedding of cells into the tubular lumen, resulting in tubular blockage and further decreasing glomerular filtration. The overall mortality rate of patients with ARF is still high despite major advances in pharmacologic therapy, intensive care, and renal replacement therapy. Therefore, a more potent therapeutic intervention for ARF to reduce mortality is imperative. Our previous study showed that endogenous bone marrow cells could contribute to the renal tubular epithelial cell population and regeneration of the renal tubular epithelium by DNA synthesis after folic acid-induced acute kidney injury, although most of the renal tubular regeneration came from indigenous cells. These results have also been supported by another study. Recently, a stem cell-based treatment strategy has started to become a realistic option to replace or rebuild damaged organs and tissues. Stem cell therapy has been successfully applied using a variety of cell types, including mesenchymal stem cells, BM cells, and human umbilical cord blood cells, to rescue organ 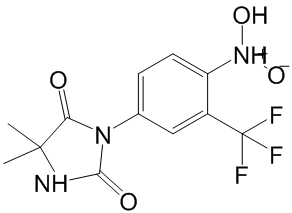 Albaspidin-AA damage in animal and human studies. On the basis of studies of allogeneic BM MSCs as a cell source for stem cell therapy for acute tubular necrosis, several studies have shown that sorted BM MSCs can rescue nonirradiated mice from acute renal tubular damage caused by toxins or ischemia; however, it is still debatable whether the beneficial effects of MSCs are primarily mediated via their differentiation into target cells, or by complex paracrine actions. BM MSCs are obtained from human bone marrow; however, aspiration of BM is an invasive procedure, and the numbers and differentiation capabilities of BM MSCs decline significantly with age. Fetal MSCs are derived from fetuses, a source associated with considerable ethical problems for human application, making these cells difficult to obtain. Compared with BM MSCs and fetal MSCs, human umbilical cord-derived mesenchymal stem cells can be separated from discarded umbilical cord, which causes no harm to the donor, and is not ethically problematic. Therefore, hUC-MSCs are a safe and accessible source for large quantities of stem cells in comparison to fetal MSCs and BM MSCs.
Albaspidin-AA damage in animal and human studies. On the basis of studies of allogeneic BM MSCs as a cell source for stem cell therapy for acute tubular necrosis, several studies have shown that sorted BM MSCs can rescue nonirradiated mice from acute renal tubular damage caused by toxins or ischemia; however, it is still debatable whether the beneficial effects of MSCs are primarily mediated via their differentiation into target cells, or by complex paracrine actions. BM MSCs are obtained from human bone marrow; however, aspiration of BM is an invasive procedure, and the numbers and differentiation capabilities of BM MSCs decline significantly with age. Fetal MSCs are derived from fetuses, a source associated with considerable ethical problems for human application, making these cells difficult to obtain. Compared with BM MSCs and fetal MSCs, human umbilical cord-derived mesenchymal stem cells can be separated from discarded umbilical cord, which causes no harm to the donor, and is not ethically problematic. Therefore, hUC-MSCs are a safe and accessible source for large quantities of stem cells in comparison to fetal MSCs and BM MSCs.
In bona fide HIF binding regions if cooperation is specific to a small number of target genes
Furthermore, the employed HIF binding data in this study is for the HIF1a subunit only, whereas transcription factor cooperativity may well apply to other HIF subunits. In fact, several reports have implicated the ETS family of transcription factors in target selection by HIF2a. We observed very similar tendencies when transcriptional activation of reporter constructs was elicited by DMOG or hypoxia treatment, additionally suggesting that, at least in our experimental conditions, the contribution of these factors could occur mainly in basal conditions, as it is unlikely that hypoxia and DMOG treatment induce completely overlapping cellular responses. Several recent reports have suggested that chromatin accessibility determines HIF1 binding, although this mechanism may not fully explain HIFs binding and target selectivity. Our results indicate that an additional layer of specificity comes from proximal co-binding of other transcription factors and HIFs to open chromatin regions, thereby Benzethonium Chloride facilitating or restricting HIF-mediated transcription. Elucidation of the underlying molecular 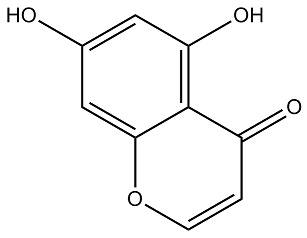 mechanisms falls outside the scope of our work, although it is tempting to speculate that transcription factors binding in proximity of HIFs may be involved in recruitment of co-activator or co-repressor proteins. Of note, a recent mammalian two-hybrid survey of protein-protein interactions for human and mouse TFs reported a physical association between HIF1A and AP-1 family member JUN, as well as the previously known interaction between CEBPB and p300. p300/CBP is a master co-activator of HIF-mediated transcription whose recruitment can also be mediated by CREB. In this regard, evidence from a synthetic transactivation screen on the EGLN1 promoter pointed to ETV4 as an additional p300-dependent coactivator of HIF-mediated transcription. Moreover, HIF1 is known to interact with Jab1/CSN5, a protein originally identified as a transcriptional coactivator for AP1. Future investigations on protein composition of HIF-bound enhancers should be pivotal in supporting this model. The associations between HIFs and AP1, CREB and CEBPs analyzed in our reporter results could be general across many HIF targets or be restricted to individual targets. To judge the generality of these results, we conducted a gene-set enrichment analysis of transcription factor targets in a sorted list of genes regulated by hypoxia. The results of this analysis showed a significant enrichment of CEBP targets among hypoxiainducible genes, suggesting that at least for this family of transcription factors, the functional association with HIFs could be relatively general. Of note, recent works have reported a direct protein-protein interaction between HIF1a and CEBPa, and have implicated CEBPa activity in regulation of the HIF target genes galectin-1 and PAI-1. Hypoxic induction of both galectin-1 and PAI-1 was found to be synergistically dependent on both HIF1a and CEBPa activity and their co-binding to the promoter region. Our results further suggest that this functional association may be general across a wider collection of HIF targets. In conclusion, the data presented herein demonstrates that integration of high-throughput chromatin immunoprecipitation and gene expression data is a successful approach to select highquality core HIF binding regions, and provides experimental proof of principle for the biological relevance of enriched transcription factor binding sites other than the HIF binding consensus in HIFmediated transcription. Specifically, our results suggest that Folinic acid calcium salt pentahydrate diverse stress-responsive transcription factors, in particular CEBPs, contribute to fine-tuning of the HIF-mediated transcriptional response. With the advent of the HIV epidemics, the disease has emerged as an important opportunistic infection in AIDS patients.
mechanisms falls outside the scope of our work, although it is tempting to speculate that transcription factors binding in proximity of HIFs may be involved in recruitment of co-activator or co-repressor proteins. Of note, a recent mammalian two-hybrid survey of protein-protein interactions for human and mouse TFs reported a physical association between HIF1A and AP-1 family member JUN, as well as the previously known interaction between CEBPB and p300. p300/CBP is a master co-activator of HIF-mediated transcription whose recruitment can also be mediated by CREB. In this regard, evidence from a synthetic transactivation screen on the EGLN1 promoter pointed to ETV4 as an additional p300-dependent coactivator of HIF-mediated transcription. Moreover, HIF1 is known to interact with Jab1/CSN5, a protein originally identified as a transcriptional coactivator for AP1. Future investigations on protein composition of HIF-bound enhancers should be pivotal in supporting this model. The associations between HIFs and AP1, CREB and CEBPs analyzed in our reporter results could be general across many HIF targets or be restricted to individual targets. To judge the generality of these results, we conducted a gene-set enrichment analysis of transcription factor targets in a sorted list of genes regulated by hypoxia. The results of this analysis showed a significant enrichment of CEBP targets among hypoxiainducible genes, suggesting that at least for this family of transcription factors, the functional association with HIFs could be relatively general. Of note, recent works have reported a direct protein-protein interaction between HIF1a and CEBPa, and have implicated CEBPa activity in regulation of the HIF target genes galectin-1 and PAI-1. Hypoxic induction of both galectin-1 and PAI-1 was found to be synergistically dependent on both HIF1a and CEBPa activity and their co-binding to the promoter region. Our results further suggest that this functional association may be general across a wider collection of HIF targets. In conclusion, the data presented herein demonstrates that integration of high-throughput chromatin immunoprecipitation and gene expression data is a successful approach to select highquality core HIF binding regions, and provides experimental proof of principle for the biological relevance of enriched transcription factor binding sites other than the HIF binding consensus in HIFmediated transcription. Specifically, our results suggest that Folinic acid calcium salt pentahydrate diverse stress-responsive transcription factors, in particular CEBPs, contribute to fine-tuning of the HIF-mediated transcriptional response. With the advent of the HIV epidemics, the disease has emerged as an important opportunistic infection in AIDS patients.
The expression of fourteen genes in individual samples by real time qPCR
The high coefficient of determination obtained confirmed the high reliability of the microarray approach. This value was even shifted up when genes displaying low expression, and therefore less reliable Ct values in the correlation were excluded. Gonad samples were collected from Pacific oysters originating from 3 different sampling sites, at the four stages of the yearly reproductive cycle of oysters. This sampling method was undertaken in order to compensate for a possible bias in Atropine sulfate transcriptome analysis due to singularity of a single population within a precise environmental context. The results were mainly obtained from studying transcriptomic profiles from site 1 individuals, and show that the transcriptome of samples collected from different geographical locations are not significantly different regarding gametogenesis. Thus, we compared the transcriptomes of gonads from site 1 and site 2 using both microarray and real time qPCR analyses. The high correlation between geographical locations confirmed that the expression profiles observed are real features of gametogenesis in oysters without significant influence of the sampling site. Our analysis provided lists of genes Catharanthine sulfate expressed in male and female gonads, genes that increase in expression along the gametogenetic cycle, genes expressed in the flagella structure of spermatozoids, genes expressed in oocytes and genes expressed by female somatic cells. Most importantly, cross-referencing these lists of genes allowed us to identify potential markers of early sex differentiation in C. gigas oyster, a singular alternative hermaphrodite mollusk. We also provided new highly valuable information on genes specifically expressed by mature spermatozoids and mature oocytes. We  initially hoped to use PCA to discriminate between male and female stage 0 oysters and to identify genes involved in early sex differentiation. However, no difference was observed between the eight stage 0 gonads analyzed. Therefore, the future sexual development of these gonads could not be predicted from this PCA analysis. Principal component analysis revealed that differences between males and females increased overtime, from stage 1 gonads to stage 3 gonads, suggesting that sex differentiation takes place sometime before. Interestingly, some of the studied stage 0 gonads were found to express male specific or female specific genes, suggesting that sex differentiation already took place within these individuals although it was not possible to sex them using histology. We did not observe mitosis and cell proliferation within these individuals by histological methods. However, we performed cytology by observing a single transverse section of the gonad collected in the middle of the organ. Heterogeneity in the development of germ cells at different levels of the gonad has been observed and this may explain that some sex specific genes are found expressed when gene expression is measured on the whole gonad. Thus, the individuals presumed to be at stage 0 according to histological characterization, may biologically correspond to early stage 1 individuals. Few differences were observed between the transcriptomic profiles of undifferentiated stage 0 and sexed stage 1 individuals. Differences seem to fit with germ cell proliferation and the onset of meiosis in stage 1. The mitosis/ meiosis and sperm/oocyte decision may take place during the same time frame in the oyster gonad. In C. elegans, fbf and gld-3 genes control both the decision to leave the mitotic cell cycle and enter meiosis and to achieve the switch from spermatogenesis to oogenesis.
initially hoped to use PCA to discriminate between male and female stage 0 oysters and to identify genes involved in early sex differentiation. However, no difference was observed between the eight stage 0 gonads analyzed. Therefore, the future sexual development of these gonads could not be predicted from this PCA analysis. Principal component analysis revealed that differences between males and females increased overtime, from stage 1 gonads to stage 3 gonads, suggesting that sex differentiation takes place sometime before. Interestingly, some of the studied stage 0 gonads were found to express male specific or female specific genes, suggesting that sex differentiation already took place within these individuals although it was not possible to sex them using histology. We did not observe mitosis and cell proliferation within these individuals by histological methods. However, we performed cytology by observing a single transverse section of the gonad collected in the middle of the organ. Heterogeneity in the development of germ cells at different levels of the gonad has been observed and this may explain that some sex specific genes are found expressed when gene expression is measured on the whole gonad. Thus, the individuals presumed to be at stage 0 according to histological characterization, may biologically correspond to early stage 1 individuals. Few differences were observed between the transcriptomic profiles of undifferentiated stage 0 and sexed stage 1 individuals. Differences seem to fit with germ cell proliferation and the onset of meiosis in stage 1. The mitosis/ meiosis and sperm/oocyte decision may take place during the same time frame in the oyster gonad. In C. elegans, fbf and gld-3 genes control both the decision to leave the mitotic cell cycle and enter meiosis and to achieve the switch from spermatogenesis to oogenesis.
Understanding of the molecular mechanisms underlying the course of a reproductive cycle of oysters by describing their gonad transcriptome
Therefore, this study was designed to provide a better to establish lists of genes of interest specific to each reproductive 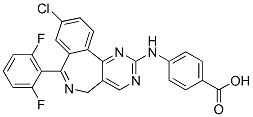 stage and sex. We employed a custom oligonucleotide microarray containing 31,918 ESTs Gomisin-D described and validated in Dheilly et al.. Our study identifies novel sex specific molecular markers and genes differentially expressed over the different stages of the gametogenesis cycle of males and females. Five hundred and eleven genes decreased in expression along the gametogenetic cycle. Numerous genes were previously identified as tissue-enriched in either the digestive gland, the mantle tissue, the visceral ganglion, hemocytes or the adductor muscle by Dheilly et al.. The method we employed did not exclude the possibility that genes that appear more expressed in early gonad developmental stages than in maturing gonads could be artifacts due to a dilution effect of genes expressed in somatic tissues, such as muscular fibers surrounding the gonadal tubules, when germ cells accumulate within the gonad area. In order to identify genes specifically expressed during early gametogenetic stages, we searched for genes significantly more expressed in immature gonads than in somatic tissues and mature gonads. Thus we compared expression data in stage 0 oyster gonads with expression data from somatic tissues previously described in Dheilly et al. in order to differentiate germline specific genes and genes somatically expressed in our gonad samples. Oyster gonad is a mixed tissue including storage tissue, smooth muscle fibers and circulating hemocytes. In order to characterize the expression of genes involved in gametogenesis in oocytes or female somatic tissue, we studied the transcriptome of oocytes collected by stripping 7 mature females and compared them to the transcriptome of the 10 stage 3 female gonad samples. Among the 2,482 genes differentially expressed in both male and female gametogenesis, 434 were significantly differentially expressed between female gonad tissue and stripped oocytes. Genes for which more transcripts were found in whole stage 3 gonads are predicted to be expressed by female somatic tissues. When more transcripts were found in stripped mature oocytes, the genes are predicted to be expressed by female germ cells. Predicted localization of gene expression is provided in file S3. Recent expression profiling studies using microarrays have provided great insight into the molecular mechanisms governing various complex physiological traits. Among those, the unprecedented amount of Butenafine hydrochloride information collected on gametogenesis, mitosis or meiosis of different eukaryotes such as yeast or nematode had a great impact on our understanding of sexual reproduction. Microarrays have also been employed successfully to better understand the cellular and molecular events of the development of reproductive tissues and of embryogenesis of cattle, mouse, rat and fish. Here, we proposed to unravel some molecular mechanisms involved in sex differentiation and gametogenesis of a peculiar alternative hermaphrodite invertebrate, the Pacific oyster Crassostrea gigas. We used an oligonucleotide microarray composed of 31,918 ESTs to characterize the transcriptome of oyster gonads at different developmental stages. The microarray employed in this study had previously been used to describe the transcriptome of various tissues of C. gigas and the results were validated by showing a significant correlation of gene expression obtained by real time qPCR and microarrays.
stage and sex. We employed a custom oligonucleotide microarray containing 31,918 ESTs Gomisin-D described and validated in Dheilly et al.. Our study identifies novel sex specific molecular markers and genes differentially expressed over the different stages of the gametogenesis cycle of males and females. Five hundred and eleven genes decreased in expression along the gametogenetic cycle. Numerous genes were previously identified as tissue-enriched in either the digestive gland, the mantle tissue, the visceral ganglion, hemocytes or the adductor muscle by Dheilly et al.. The method we employed did not exclude the possibility that genes that appear more expressed in early gonad developmental stages than in maturing gonads could be artifacts due to a dilution effect of genes expressed in somatic tissues, such as muscular fibers surrounding the gonadal tubules, when germ cells accumulate within the gonad area. In order to identify genes specifically expressed during early gametogenetic stages, we searched for genes significantly more expressed in immature gonads than in somatic tissues and mature gonads. Thus we compared expression data in stage 0 oyster gonads with expression data from somatic tissues previously described in Dheilly et al. in order to differentiate germline specific genes and genes somatically expressed in our gonad samples. Oyster gonad is a mixed tissue including storage tissue, smooth muscle fibers and circulating hemocytes. In order to characterize the expression of genes involved in gametogenesis in oocytes or female somatic tissue, we studied the transcriptome of oocytes collected by stripping 7 mature females and compared them to the transcriptome of the 10 stage 3 female gonad samples. Among the 2,482 genes differentially expressed in both male and female gametogenesis, 434 were significantly differentially expressed between female gonad tissue and stripped oocytes. Genes for which more transcripts were found in whole stage 3 gonads are predicted to be expressed by female somatic tissues. When more transcripts were found in stripped mature oocytes, the genes are predicted to be expressed by female germ cells. Predicted localization of gene expression is provided in file S3. Recent expression profiling studies using microarrays have provided great insight into the molecular mechanisms governing various complex physiological traits. Among those, the unprecedented amount of Butenafine hydrochloride information collected on gametogenesis, mitosis or meiosis of different eukaryotes such as yeast or nematode had a great impact on our understanding of sexual reproduction. Microarrays have also been employed successfully to better understand the cellular and molecular events of the development of reproductive tissues and of embryogenesis of cattle, mouse, rat and fish. Here, we proposed to unravel some molecular mechanisms involved in sex differentiation and gametogenesis of a peculiar alternative hermaphrodite invertebrate, the Pacific oyster Crassostrea gigas. We used an oligonucleotide microarray composed of 31,918 ESTs to characterize the transcriptome of oyster gonads at different developmental stages. The microarray employed in this study had previously been used to describe the transcriptome of various tissues of C. gigas and the results were validated by showing a significant correlation of gene expression obtained by real time qPCR and microarrays.