The mutational analysis from the static structure normally ignores short or long range conformational changes and they do not include the dynamic effects caused by thermal motions. The molecular dynamics simulations and molecular mechanics-Poisson-Boltzmann surface area calculations on the problem of imatinib resistance by various BCR-ABL mutations has been studied by Lee et al.,. Computational simulations can provide atomic level description of structural details, energy landscape, dynamic behaviours, and other properties which are difficult to be obtained from the experimental studies. Here, we report the MD simulations, solvated interaction energies free energy calculations of ponatinib with native and mutants of BCR-ABL kinase. We have also calculated the contributions from individual amino acid BEZ235 residues in the active site of all complexes to provide the molecular basis for inhibition. To our knowledge these Reversine studies have not been carried out before and our results provide detailed information about the molecular mechanisms of inhibition of native and various mutant BCR-ABL tyrosine kinases when bound to ponatinib. The native and mutant ABL kinase �C ponatinib complexes with explicit water molecules and sodium ions for charge neutralization were subjected to 25 ns MD simulations. The fourteen BCR-ABL mutants studied in this work collectively represent more than 95% of clinically observed mutations that are imatinib resistant. With the exception of T315I, most BCR-ABL mutations are inhibited by dasatinib and nilotinib. Ponatinib inhibits native and all mutant ABL kinases with high affinity, although some mutants have slightly greater inhibition than the others. The ATP competitive inhibitors of ABL kinase are classified into DFG-in or DFG-out classes depending on their binding interactions with kinase domain. Ponatinib binds to ABL kinase domain with a DFG-out conformation and serves to distribute binding energy over a wide range of amino acid residues in the active site as shown in Figure 1. The presence of such optimized and distributed binding interactions has the potential to allow ponatinib to withstand modest reduction in potency caused by single mutation. For our convenience; we grouped these mutations by the region of their location in ABL kinase structure. These regions include the P-loop mutants M244V, G250E, Q252H, Y253F, Y253H, E255K, and E255V; gatekeeper residue mutants T315A and T315I; hinge region mutants F317L and F317V; activation loop mutant H396P and other mutants M351T and F359V. The location of mutations in BCR-ABL kinase is shown in Figure 2. In the ABL kinase, amino acid residues Tyr253, Thr315, Phe317 and Phe359 are located in close contact with ponatinib and therefore affect the binding and activity of inhibitor. The Ploop mutant residues Gly250, Gln252 and Glu255 are not in direct contact with ponatinib, but share non-bonding interactions with inhibitor. The rest of the mutations Met244, Met351 and His396 are located away from inhibitor binding site, but intriguingly display ponatinib based inhibition. SIE calculations from MD trajectories measure the free energy of complex formation. Table 1 shows the calculated free energies for native and 14 mutant BCR-ABL �C ponatinib complexes. The intermolecular vdW, intermolecular coulomb and change in surface area are shown in Table 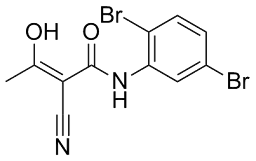 1. This table indicates that IC50 values vary from 0.5 nM to 36 nM and SIE values calculated from this work are in the range 210.03 kcal/mol to 210.67 kcal/mol. Though there is no direct correlation between IC50 and SIE values, it can be observed that their respective values lie within a narrow range. Many patients eventually developed imatinib resistance, usually associated with above mentioned mutations in ABL kinase domain that either directly or indirectly effects the binding affinity of imatinib to ABL. The most common gatekeeper residue mutation T315I that accounts for 15�C20% of clinically observed mutations is completely resistant to imatinib, nilotinib and dasatinib. Native and T315I BCR-ABL kinases complexed with dasatinib are subjected to 25 ns of MD simulations and SIE binding free energies are calculated. The analysis of dasatinib complexed with native and T315I mutant BCR-ABL kinases revealed that native complex has relatively higher SIE free energy than when complexed with T315I that signifies the greater affinity of dasatinib for native compared to mutant BCR-ABL kinase.
1. This table indicates that IC50 values vary from 0.5 nM to 36 nM and SIE values calculated from this work are in the range 210.03 kcal/mol to 210.67 kcal/mol. Though there is no direct correlation between IC50 and SIE values, it can be observed that their respective values lie within a narrow range. Many patients eventually developed imatinib resistance, usually associated with above mentioned mutations in ABL kinase domain that either directly or indirectly effects the binding affinity of imatinib to ABL. The most common gatekeeper residue mutation T315I that accounts for 15�C20% of clinically observed mutations is completely resistant to imatinib, nilotinib and dasatinib. Native and T315I BCR-ABL kinases complexed with dasatinib are subjected to 25 ns of MD simulations and SIE binding free energies are calculated. The analysis of dasatinib complexed with native and T315I mutant BCR-ABL kinases revealed that native complex has relatively higher SIE free energy than when complexed with T315I that signifies the greater affinity of dasatinib for native compared to mutant BCR-ABL kinase.
Monthly Archives: July 2019
the high accumulation of p53 renders the cells highly susceptible to p53 reactivation and more sensitive to apoptosis
From a therapeutic perspective, it is interesting that ISA27 in combination with the conventional chemotherapy drug TMZ inhibited U87MG cell growth. This combination worked in a synergistic manner as confirmed by isobolographic GDC-0199 Bcl-2 inhibitor analysis. This result suggests the possibility of lowering the dose of TMZ used in the treatment of GBM. In conclusion, our data show that ISA27 disrupts the MDM2p53 interaction and releases the powerful antitumor capacities of p53 in GBM cells. The use of this MDM2 inhibitor could offer a novel therapy for the treatment of GBM patients by inhibiting tumor growth. Proteases catalyze the hydrolysis of peptide bonds in proteins and are involved in digestive as well as regulatory processes. In the human genome, approximately 2% of the genes code for proteases. While most proteases are soluble, a small fraction is membrane-embedded. These intramembrane proteases differ from soluble proteases in a variety of aspects: They are composed of a number of transmembrane domains which harbor the catalytic residues with their active sites buried several A ? into the membrane. Their substrates are transmembrane proteins that reside inside the membrane in a dormant form. Upon cleavage, most substrates release a soluble part into the cytosol or extracellular space. It is therefore not surprising that intramembrane proteases are involved in various signaling pathways. There are three families of intramembrane proteases, classified according to their catalytic mechanism: intramembrane metalloproteases, intramembrane aspartic proteases, and intramembrane serine proteases. The latter belong to the family of rhomboid proteins, containing active intramembrane proteases and inactive homologs. Rhomboids are found in all kingdoms of life, but are functionally diverse. They take part in various distinct cellular processes such as the EGFR-signaling pathway in the fruit fly Drosophila melanogaster, quorum sensing in the Gram negative bacterium Providencia stuartii and host cell infection by apicomplexan parasites. Structurally, rhomboids are the best characterized intramembrane proteases. Several different crystal forms of the E. coli rhomboid GlpG have provided insight into the mechanism of intramembrane proteolysis. However, a detailed picture of the rhomboid-substrate interaction is not available. As an alternative, crystal structures of covalent inhibitors bound to GlpG have revealed which areas and residues may play a role in primed and non-primed site interaction, and oxyanion stabilization. The availability of inhibitors is also important for future functional studies. Moreover, potent and BAY 73-4506 selective inhibitors may serve as lead structures for future drug design. Up to date, rhomboid inhibitors have been reported based on three distinct scaffolds: 4-chloro-isocoumarins, fluorophosphonates, and N-sulfonylated beta-lactams. However, these are not selective enough to inhibit only rhomboids within the entire proteome. In addition, these inhibitors are also not promiscuous enough to inhibit rhomboids from different organisms  equally well. Therefore, it is still of great interest to find new types of inhibitors. In order to facilitate this search, various screening methods have been employed so far. All of these have relied on monitoring the cleavage of a substrate through gel-based, FRET or MALDI mass spectrometry techniques. However, a limitation of these methods is the availability of a matching protein or polypeptide substrate. Rhomboids from one species may cleave substrates from another species, but this is not a general rule. We therefore reasoned that it would be beneficial to develop an inhibitor assay for rhomboid proteases that does not rely on a substrate at all. A few years ago Cravatt and co-workers developed a highthroughput inhibitor screening method that uses fluorescent activity-based probes. ABPs are small molecules that covalently bind to the active form of an enzyme, but not to an inactivated or zymogen form. ABPs generally consist of a tag, a spacer and an electrophilic group that traps an active site nucleophile. The binding event can be detected by a variety of techniques, such as gel-scanning, biotin blot or fluorescent microscopy, depending on the tagging moiety. When appended to a fluorescent dye, the binding of an ABP can be detected by fluorescence polarization. This so-called fluorescence polarization activity-based protein profiling has been used in inhibitor high-throughput screens for a variety of poorly characterized enzymes.
equally well. Therefore, it is still of great interest to find new types of inhibitors. In order to facilitate this search, various screening methods have been employed so far. All of these have relied on monitoring the cleavage of a substrate through gel-based, FRET or MALDI mass spectrometry techniques. However, a limitation of these methods is the availability of a matching protein or polypeptide substrate. Rhomboids from one species may cleave substrates from another species, but this is not a general rule. We therefore reasoned that it would be beneficial to develop an inhibitor assay for rhomboid proteases that does not rely on a substrate at all. A few years ago Cravatt and co-workers developed a highthroughput inhibitor screening method that uses fluorescent activity-based probes. ABPs are small molecules that covalently bind to the active form of an enzyme, but not to an inactivated or zymogen form. ABPs generally consist of a tag, a spacer and an electrophilic group that traps an active site nucleophile. The binding event can be detected by a variety of techniques, such as gel-scanning, biotin blot or fluorescent microscopy, depending on the tagging moiety. When appended to a fluorescent dye, the binding of an ABP can be detected by fluorescence polarization. This so-called fluorescence polarization activity-based protein profiling has been used in inhibitor high-throughput screens for a variety of poorly characterized enzymes.
ASM is bound to intralysosomal membranes and thereby protected against its own proteolytic inactivation
However, studies using cells derived from Niemann-Pick disease patients or from ASM knock-out mice revealed that the deficiency of this enzyme might also have beneficial consequences, including anti-apoptotic and cytoprotective effcts. In fact, there is increasing evidence that ASM activation and MK-4827 ceramide accumulation play a central role in the development of common human diseases. Reports have been published of aberrant activation of ASM and/or altered levels of ceramide, for instance, for several psychiatric and neurological disorders such as major depression, morphine antinociceptive tolerance, Alzheimer’s disease, spinal cord injuryand seizure disorder. Therefore, ASM inhibitors hold promise for a number of new clinical therapies and might be used to prevent apoptosis and other negative effects occurring in different disease states such as in ischemia, stroke, Alzheimer’s dementia, Parkinson’s disease, Huntington’s chorea, and of certain infections, in endotoxemia, and in atherosclerosis, and for the therapy of major depressive disorder. Currently, only few examples of inhibitors directly interacting with ASM are known. These substances include physiological inhibitors of ASM such as phosphatidyl-myoinositol-3,4,5-trisphosphate, L-a-phosphatidyl-D-myo-inositol3,5-bisphosphateand non-natural direct inhibitors of ASM, such as SMA-7and AD2765. A high throughput screening for direct ASM inhibitors was unsuccessful in finding lead structures. The rational development of compounds that block ASM by direct interaction with the enzyme is difficult, since the crystal structure of the enzyme is not yet available. On the other hand, it has been known since the 1970s that some weak organic bases have the potential to reduce the activity of ASM. Weak bases, such as desipramine 168, strongly accumulate in acidic intracellular compartments like lysosomes, a phenomenon called ‘acid trapping’  and in the case of lysosomes ‘lysosomotropism’. This accumulation of desipramine 168 results in detachment of the ASM from the inner lysosomal membraneand its subsequent inactivation, probably by proteolytic degradation. Weak bases, therefore, do not directly inhibit ASM, but result in a functional inhibition of ASM. We have thus proposed the acronym FIASMA for Functional Inhibitor of Acid SphingoMyelinAse. According to this model, functional inhibition of ASM requires high lysosomal concentrations of a weak basic drug. Previously, we have shown that functional inhibition of ASM is related to high pKa- and high logP-values and have characterized several new FIASMAs, including the antidepressant drugs doxepine 63, fluoxetine 104, maprotilin 109, nortriptyline 114, paroxetine 118 and sertraline 124. The aims of the present study wereto identify more FIASMAs,to further improve the in silico prediction of functional ASM inhibition by developing compact and easily-interpretable models with high internal Talazoparib consistency,to investigate the relationship between permeation of the blood-brain barrier and functional inhibition of ASM andto study the distribution of FIASMAs across different classes of drugs licensed for medical use in humans. Using the present knowledge about lysosomal accumulation and drug-membrane interaction as prerequisite to functional inhibition of ASM, we developed the following hypotheses: FIASMAshave shared structural and physicochemical properties allowing high lysosomal drug concentrations, compatible with partitioning into the inner surface of the lysosomal membrane and alteration of electrostatic membrane properties. We hypothesize that FIASMAswill be found in different therapeutic drug classes andbelong to diverse structural drug classes.
and in the case of lysosomes ‘lysosomotropism’. This accumulation of desipramine 168 results in detachment of the ASM from the inner lysosomal membraneand its subsequent inactivation, probably by proteolytic degradation. Weak bases, therefore, do not directly inhibit ASM, but result in a functional inhibition of ASM. We have thus proposed the acronym FIASMA for Functional Inhibitor of Acid SphingoMyelinAse. According to this model, functional inhibition of ASM requires high lysosomal concentrations of a weak basic drug. Previously, we have shown that functional inhibition of ASM is related to high pKa- and high logP-values and have characterized several new FIASMAs, including the antidepressant drugs doxepine 63, fluoxetine 104, maprotilin 109, nortriptyline 114, paroxetine 118 and sertraline 124. The aims of the present study wereto identify more FIASMAs,to further improve the in silico prediction of functional ASM inhibition by developing compact and easily-interpretable models with high internal Talazoparib consistency,to investigate the relationship between permeation of the blood-brain barrier and functional inhibition of ASM andto study the distribution of FIASMAs across different classes of drugs licensed for medical use in humans. Using the present knowledge about lysosomal accumulation and drug-membrane interaction as prerequisite to functional inhibition of ASM, we developed the following hypotheses: FIASMAshave shared structural and physicochemical properties allowing high lysosomal drug concentrations, compatible with partitioning into the inner surface of the lysosomal membrane and alteration of electrostatic membrane properties. We hypothesize that FIASMAswill be found in different therapeutic drug classes andbelong to diverse structural drug classes.
Internalized transferrin regionsand MHCI was observed in some recycling tubules as described reported
Future development of additional SK2-selective inhibitors may provide more effective anticancer agents than SK1/2-dual or SK1-selective inhibitor. Cells use a variety of means to internalize extracellular material and plasma membraneby the general process of endocytosis. All cells use this process to deliver extracellular nutrients into the cell interior, recycle PM to other regions of the cell surface, and to degrade PM proteins and lipids. Clathrin-dependent endocytosisis an efficient and selective process whereby PM proteins containing specific cytoplasmic sorting sequences are gathered by adaptor proteins into clathrin-coated pits, and then are severed from the PM with the assistance of the dynamin 2 GTPase. CDE is widely studied, whereas much less is known about clathrinindependent endocytosisalthough there is evidence of CIE in many cell types and multiple pathways have been characterized. CIE includes modes of internalization for glycolipidbinding toxins such as shiga and cholera toxin, for GPIanchored proteins, for the EGF receptor under certain conditions, and for a number of endogenous PM proteins Doxorubicin involved in immune function, nutrient uptake, and cellcell and cell-matrix interactions. There is a growing list of membrane proteins entering mammalian cells by CIE and there is now good evidence that CIE exists in lower eukaryotes. The identification of selective inhibitors of CDE and CIE would greatly enhance the characterization of specific physiological functions of these endocytic processes. Many approaches have been taken to inhibit  CDE. The expression of mutants of proteins involved in the clathrin machinery, such as Dynamin2-K44A, the carboxy terminus of AP180, and clathrin hubs, has proven quite effective. More recently siRNA-mediated depletion of the clathrin heavy chain, subunits of the AP2 adaptor, and dynamin 2have abolished CDE in cells. The drawback of these genetic approaches is that they require days to take effect and may lead to many indirect effects or compensatory cellular responses that make interpretation of the findings sometimes difficult. Use of a number of acute cellular treatments including cytosol acidification and hypotonic treatment can be effective at blocking endocytosis of CDE cargobut these treatments are non-specific and may also affect CIE. Recently, new compounds that selectively target proteins involved in CDE have been identified with the promise that these could be used to acutely inhibit this process. These include compounds that specifically target dynamin such as dynasoreand the dynoles. Since dynamin is required for all forms of CDE and is used in some forms of CIE, a compound that selectively targets clathrin was developed by Haucke and colleagues. This compound, named pitstop 2, was designed and shown to bind to and block interactions between the amino terminal domain of clathrin heavy chain and amphiphysin, one of many proteins shown to bind to this domain of clathrin. In cells, pitstop 2 was shown to inhibit endocytosis of transferrin receptor, a CDE cargo TWS119 protein, but not affect endocytosis of shiga toxin, which enters cells independently of clathrin. We attempted to use pitstop to acutely block CDE in order to examine effects of blocking CDE on subsequent trafficking of endocytosed CIE cargo proteins. Surprisingly, we found that pitstop 2 potently blocks endocytosis of endogenous proteins normally entering cells by CIE. CDE and CIE can be observed in HeLa cells by monitoring endocytosis of labeled transferrin and an antibody to the Major Histocompatibility Complex Class I protein, respectively.
CDE. The expression of mutants of proteins involved in the clathrin machinery, such as Dynamin2-K44A, the carboxy terminus of AP180, and clathrin hubs, has proven quite effective. More recently siRNA-mediated depletion of the clathrin heavy chain, subunits of the AP2 adaptor, and dynamin 2have abolished CDE in cells. The drawback of these genetic approaches is that they require days to take effect and may lead to many indirect effects or compensatory cellular responses that make interpretation of the findings sometimes difficult. Use of a number of acute cellular treatments including cytosol acidification and hypotonic treatment can be effective at blocking endocytosis of CDE cargobut these treatments are non-specific and may also affect CIE. Recently, new compounds that selectively target proteins involved in CDE have been identified with the promise that these could be used to acutely inhibit this process. These include compounds that specifically target dynamin such as dynasoreand the dynoles. Since dynamin is required for all forms of CDE and is used in some forms of CIE, a compound that selectively targets clathrin was developed by Haucke and colleagues. This compound, named pitstop 2, was designed and shown to bind to and block interactions between the amino terminal domain of clathrin heavy chain and amphiphysin, one of many proteins shown to bind to this domain of clathrin. In cells, pitstop 2 was shown to inhibit endocytosis of transferrin receptor, a CDE cargo TWS119 protein, but not affect endocytosis of shiga toxin, which enters cells independently of clathrin. We attempted to use pitstop to acutely block CDE in order to examine effects of blocking CDE on subsequent trafficking of endocytosed CIE cargo proteins. Surprisingly, we found that pitstop 2 potently blocks endocytosis of endogenous proteins normally entering cells by CIE. CDE and CIE can be observed in HeLa cells by monitoring endocytosis of labeled transferrin and an antibody to the Major Histocompatibility Complex Class I protein, respectively.
The final consequences were a decrease in phosphorylation at microtubules
lt of an inhibition of AKT by reducing its phosphorylation at Ser473 and Thr 308, which regulates its activity positively. AKT inhibition may also contribute to the thiamet-G�Cinduced decrease of tau phosphorylation at Thr212 and Ser214, because these two sites are also substrates of AKT. AKT phosphorylation was mainly catalyzed by the mTORC2 complex and PI3K�Cphosphoinositide-dependent protein kinase-1. Because we did not observe the corresponding decrease in PI3K, we speculate that the reduction/elimination of phosphorylation of AKT and GSK-3b after RWJ 64809 thiamet-G treatment may result from elevation of O-GlcNAcylation of AKT, PDK1 and/or mTOR. Alternatively, it could be off-target effect of the  inhibitor when used at high doses. Phosphorylation of these kinases has been reported to be regulated negatively by O-GlcNAcylation. It is worth noting that different effects of OGA inhibition on phosphorylation of AKT and GSK-3 have been reported. Elevation of O-GlcNAcylation in skeletal muscles after OGA inhibition using another inhibitor, PUGNAc, does not significantly alter insulin-stimulated phosphorylation of AKT or GSK-3. In differentiated 3T3-L1 adipocytes, two different OGA inhibitors have been found to increase O-GlcNAc levels but not alter insulin-stimulated phosphorylation of AKT nor induce insulin resistance either. Therefore, it remains somewhat unclear as to the effects of OGA inhibition on alteration of GSK3b levels and AKT activity; the effects observed here could stem from high-dose inhibition of OGA, or alternatively from off-target effects of using the inhibitor at a high dose. Tau is abnormally hyperphosphorylated and aggregated in AD and other tauopathies. Previous studies from our and other groups have demonstrated differential roles of tau phosphorylation at various phosphorylation sites. A quantitative in vitro study demonstrated that phosphorylation of tau at Ser262, Thr231, and Ser235 inhibits its binding to microtubules by,35%,,25%, and,10%, respectively. In vitro kinetic studies of the binding between hyperphosphorylated tau and normal tau suggest that Ser199/Ser202/Thr205, Thr212, Thr231/Ser235, Ser262/ Ser356 and Ser422 are among the critical phosphorylation sites that convert tau to an inhibitory molecule that sequesters normal microtubule-associated proteins from microtubules. Further phosphorylation at Thr231, Ser396, and Ser422 promotes selfaggregation of tau into filaments. It is obvious that tau phosphorylation at various sites impacts tau activity and aggregation collectively. Our recent study has demonstrated that tau phosphorylation at the proline-rich region, which is located upstream of the microtubule-binding domains, inhibits its microtubule Niraparib assembly activity moderately and promotes its selfaggregation slightly. Tau phosphorylation at the C-terminal tail region increases its activity and promotes its self-aggregation markedly. Tau phosphorylation at both of these regions plus the microtubule-binding region nearly diminishes its activity and disrupts microtubules. Therefore, the overall impacts of thiamet-G on tau need to be further verified by its functional studies, and testing the effects of thiamet-G on cognitive function in mouse AD models, especially using different doses, becomes urgent before considering it to be a therapeutic agent for treating AD. In conclusion, thiamet-G is a specific OGA inhibitor and is very effective in elevating protein O-GlcNAcylation level in the mammalian brain. Because thiamet-G not only directly modulated tau phosphorylation inversely, but also stimulated GSK-3b activity likely via inhibition of AKT.
inhibitor when used at high doses. Phosphorylation of these kinases has been reported to be regulated negatively by O-GlcNAcylation. It is worth noting that different effects of OGA inhibition on phosphorylation of AKT and GSK-3 have been reported. Elevation of O-GlcNAcylation in skeletal muscles after OGA inhibition using another inhibitor, PUGNAc, does not significantly alter insulin-stimulated phosphorylation of AKT or GSK-3. In differentiated 3T3-L1 adipocytes, two different OGA inhibitors have been found to increase O-GlcNAc levels but not alter insulin-stimulated phosphorylation of AKT nor induce insulin resistance either. Therefore, it remains somewhat unclear as to the effects of OGA inhibition on alteration of GSK3b levels and AKT activity; the effects observed here could stem from high-dose inhibition of OGA, or alternatively from off-target effects of using the inhibitor at a high dose. Tau is abnormally hyperphosphorylated and aggregated in AD and other tauopathies. Previous studies from our and other groups have demonstrated differential roles of tau phosphorylation at various phosphorylation sites. A quantitative in vitro study demonstrated that phosphorylation of tau at Ser262, Thr231, and Ser235 inhibits its binding to microtubules by,35%,,25%, and,10%, respectively. In vitro kinetic studies of the binding between hyperphosphorylated tau and normal tau suggest that Ser199/Ser202/Thr205, Thr212, Thr231/Ser235, Ser262/ Ser356 and Ser422 are among the critical phosphorylation sites that convert tau to an inhibitory molecule that sequesters normal microtubule-associated proteins from microtubules. Further phosphorylation at Thr231, Ser396, and Ser422 promotes selfaggregation of tau into filaments. It is obvious that tau phosphorylation at various sites impacts tau activity and aggregation collectively. Our recent study has demonstrated that tau phosphorylation at the proline-rich region, which is located upstream of the microtubule-binding domains, inhibits its microtubule Niraparib assembly activity moderately and promotes its selfaggregation slightly. Tau phosphorylation at the C-terminal tail region increases its activity and promotes its self-aggregation markedly. Tau phosphorylation at both of these regions plus the microtubule-binding region nearly diminishes its activity and disrupts microtubules. Therefore, the overall impacts of thiamet-G on tau need to be further verified by its functional studies, and testing the effects of thiamet-G on cognitive function in mouse AD models, especially using different doses, becomes urgent before considering it to be a therapeutic agent for treating AD. In conclusion, thiamet-G is a specific OGA inhibitor and is very effective in elevating protein O-GlcNAcylation level in the mammalian brain. Because thiamet-G not only directly modulated tau phosphorylation inversely, but also stimulated GSK-3b activity likely via inhibition of AKT.