Using HCC and adjacent normal liver samples we investigated the gene and sCNV changes associated with tumorigenesis by comprehensively discovering the significant Chlorhexidine hydrochloride relationships within and between DNA copy number variation, global gene expression in TU and AN tissue and patient survival. Analysis of these data revealed the appearance of highly significant network changes as shown by gene pairs differentially correlated between AN and TU tissue. Interestingly this process largely consisted of loss of correlation in the TU samples consistent with disruption of normal networks. A subset of the changes observed involved gain of correlation in TU indicating the formation of new networks in some cases. Consistent with the view that loss of connectivity may represent loss of functionality and gain of connectivity may be gain of functionality, the LOC subset of genes was enriched for genes involved in normal liver function that might be expected to be largely extraneous to the needs of the tumor, whereas the GOC subset of genes is enriched in the essential tumor function of cell cycle. This appearance of loss and gain of connectivity during tumorigenesis may therefore be analogous to the long established concepts of tumor suppressers and oncogenes. Given the relative abundance of  LOC versus GOC events this implies that tumorigenesis in HCC at least is to a large degree one of disruption of tumor suppressing normal networks. Although smaller in number the GOC genes likely represent functions selected as important for disease progression and as such may be important points of intervention. Genes in TU were found to be strongly associated in cis and in trans with sCNV frequently involving large chromosomal regions. Within the architecture of sCNV-to-gene associations a number of hotspots were found where many more genes were associated with a particular marker than would be expected by chance. Additionally the genes associated with the hotspots were highly overlapping suggesting that multiple different loci may coordinately regulate a core subset of genes. The finding of hotspots in cancer data may not be unique to HCC in that similar associations, even involving the same genes and sCNV loci were found in an independent collection of cancer cell lines. Given the common architecture of sCNV across many tumor types, the cis and trans Lomitapide Mesylate correlations documented here may therefore be relevant to a broad range of diseases. The differentially connected genes between AN and TU tissue were also significantly enriched for association to sCNV markers in TU suggesting that the network transitions and associated functional changes may be mediated by somatic sCNV. A surprising finding in this study was that at the same FDR, three times as many genes predictive of survival were found in AN than in TU tissue. Furthermore, although the AN-survival and TU-survival genes overlapped more than would be expected by chance, the majority of genes in each case were not predictive in the other tissue. A direct connection between the altered predictive value of the genes in AN and TU was found by association to sCNV markers where AN-survival genes were preferentially associated with sCNV markers in TU that were not predictive, and TU-survival genes were enriched for association to predictive sCNV markers. It therefore seems that the sCNV in tumors may be sufficient to explain the transformation of the predictive value of genes in AN versus TU. To directly address the hypothesis of whether the pre-existing state of genes, we measured the transcriptional signature of a treatment that promotes HCC tumorigenesis.
LOC versus GOC events this implies that tumorigenesis in HCC at least is to a large degree one of disruption of tumor suppressing normal networks. Although smaller in number the GOC genes likely represent functions selected as important for disease progression and as such may be important points of intervention. Genes in TU were found to be strongly associated in cis and in trans with sCNV frequently involving large chromosomal regions. Within the architecture of sCNV-to-gene associations a number of hotspots were found where many more genes were associated with a particular marker than would be expected by chance. Additionally the genes associated with the hotspots were highly overlapping suggesting that multiple different loci may coordinately regulate a core subset of genes. The finding of hotspots in cancer data may not be unique to HCC in that similar associations, even involving the same genes and sCNV loci were found in an independent collection of cancer cell lines. Given the common architecture of sCNV across many tumor types, the cis and trans Lomitapide Mesylate correlations documented here may therefore be relevant to a broad range of diseases. The differentially connected genes between AN and TU tissue were also significantly enriched for association to sCNV markers in TU suggesting that the network transitions and associated functional changes may be mediated by somatic sCNV. A surprising finding in this study was that at the same FDR, three times as many genes predictive of survival were found in AN than in TU tissue. Furthermore, although the AN-survival and TU-survival genes overlapped more than would be expected by chance, the majority of genes in each case were not predictive in the other tissue. A direct connection between the altered predictive value of the genes in AN and TU was found by association to sCNV markers where AN-survival genes were preferentially associated with sCNV markers in TU that were not predictive, and TU-survival genes were enriched for association to predictive sCNV markers. It therefore seems that the sCNV in tumors may be sufficient to explain the transformation of the predictive value of genes in AN versus TU. To directly address the hypothesis of whether the pre-existing state of genes, we measured the transcriptional signature of a treatment that promotes HCC tumorigenesis.
Monthly Archives: June 2019
Expression in mouse liver produced a gene signature prior to the appearance of tumors
Significantly enriched in the human AN-survival genes, in genes that participate in human HCC network changes, and in genes associated in human HCC with sCNV. This is directly supports the hypothesis that the pre-tumor state, as measured by the AN tissue, was a significant determinant of the large scale network transformations required to produce HCC tumorigenesis. There are a number of interesting ideas that derive from this hypothesis. One is that MET overexpression causes increased tumorigenesis by altering the genes that participate in that transition, or in other words the starting state of these genes is causally related to the probability of the future transformation occurring. Similarly then, the starting state of the AN-survival genes may be causal for the probability of network transformations involving them in human HCC. The TU-survival genes in an analogous manner may also be causally related to the probability of future network evolutions relevant to disease progression. This further suggests, that as in the MET case, manipulation of the relevant genes will alter the probability of HCC network changes and tumor evolution occurring. The finding that AN-survival genes for the most part  lose their predictive value in tumor is interesting. By implication once the network transformation has occurred those genes and their associated functions were generally no longer rate limiting. This is apparently largely true for both disruption of normal networks and creation of new networks. From a perspective of targeting tumors, disruption of normal networks may be hard to reverse in practice. However the creation of new networks may represent functions that the tumor has gained or emphasized relative to the tissue from which it was derived. As such these functions may make desirable targets in that the tumors have selected for them and the selection process may relate to survival. Targeting these new networks may therefore disrupt essential tumor specific functions. Finally, TU-survival genes may also represent an opportunity for intervention in that as described above they may causally relate to the probability of future disease progression. Investigation of coexpression networks highlighted 4 co-expression modules that were enriched for TU-survival genes. Two of the 4 modules were strongly linked to Gomisin-D ribosomes and ribosome biogenesis, which have been linked to aggressive disease in other tumor types and individual components when either over or under expressed promote tumorigenesis. Myc has been shown to alter a number of ribosome components and in turn can be regulated by them and was found here to be in the same co-expression network. This suggests that altered translation maybe a significant factor in HCC disease progression. A third co-expression module was unusually found to be enriched for both AN and TU-survival genes and centered around metabolism and the mitochondrion. Although it is speculation it is tempting to suggest that this group of genes may represent the molecular equivalent of the Orbifloxacin epidemiological observation that obesity is a risk factor for susceptibility to HCC and survival after diagnosis. Interestingly it was recently suggested that switching to a low fat diet alters the course of disease in mouse models. Gliomas are the most common primary brain tumors, characterized by the infiltration of neighboring brain structures and robust expansion during progression to a glioblastoma multiforme. Gliomas are frequently characterized by dysregulated signaling downstream of growth factor receptors such as EGFR, PDGFR, and IGFR, and elevated production of their corresponding ligands.
lose their predictive value in tumor is interesting. By implication once the network transformation has occurred those genes and their associated functions were generally no longer rate limiting. This is apparently largely true for both disruption of normal networks and creation of new networks. From a perspective of targeting tumors, disruption of normal networks may be hard to reverse in practice. However the creation of new networks may represent functions that the tumor has gained or emphasized relative to the tissue from which it was derived. As such these functions may make desirable targets in that the tumors have selected for them and the selection process may relate to survival. Targeting these new networks may therefore disrupt essential tumor specific functions. Finally, TU-survival genes may also represent an opportunity for intervention in that as described above they may causally relate to the probability of future disease progression. Investigation of coexpression networks highlighted 4 co-expression modules that were enriched for TU-survival genes. Two of the 4 modules were strongly linked to Gomisin-D ribosomes and ribosome biogenesis, which have been linked to aggressive disease in other tumor types and individual components when either over or under expressed promote tumorigenesis. Myc has been shown to alter a number of ribosome components and in turn can be regulated by them and was found here to be in the same co-expression network. This suggests that altered translation maybe a significant factor in HCC disease progression. A third co-expression module was unusually found to be enriched for both AN and TU-survival genes and centered around metabolism and the mitochondrion. Although it is speculation it is tempting to suggest that this group of genes may represent the molecular equivalent of the Orbifloxacin epidemiological observation that obesity is a risk factor for susceptibility to HCC and survival after diagnosis. Interestingly it was recently suggested that switching to a low fat diet alters the course of disease in mouse models. Gliomas are the most common primary brain tumors, characterized by the infiltration of neighboring brain structures and robust expansion during progression to a glioblastoma multiforme. Gliomas are frequently characterized by dysregulated signaling downstream of growth factor receptors such as EGFR, PDGFR, and IGFR, and elevated production of their corresponding ligands.
Leads to enhanced bAPP degradation and reduced Ab peptide secretion has been suggested
While we cannot exclude the possibility that glial cells are providing some neuroprotective ‘shielding’, both neuronal and glial cells release cytokines when exposed to Ab42 that, in turn, activate more microglia and astrocytes that reinforce pathogenic signaling. NPD1 is anti-inflammatory and promotes inflammatory resolution. In HNG cell models of Ab42 toxicity, microarray analysis and Western blot analysis Chlorhexidine hydrochloride revealed down-regulation of pro-inflammatory genes, suggesting NPD1’s anti-inflammatory bioactivity targets, in part, this gene family. These effects are persistent, as shown by time-course Western blot analysis in which protein expression was examined up to 12 h after treatment by Ab42 and NPD1. Although counteracting Ab42-induced neurotoxicity is a promising strategy for AD treatment, curbing excessive Ab42 release during neurodegeneration is also desirable. DHA could lower Ab42 load in the CNS by stimulating non-amyloidogenic bAPP processing, reducing PS1 expression, or by increasing the expression of the sortilin receptor, SorLA/LR11. In contrast to a previous Lomitapide Mesylate report by Green et al. that suggested that Ab peptide reductions in whole brain homogenates of 3xTg AD after dietary supplementation of DHA were the result of decreases in the steady state levels of PS1, our experiments in primary HNG cells showed no 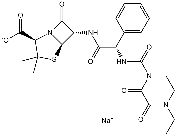 effects of NPD1 on PS1 levels, but a significant increase in ADAM10 coupled to a decrease in BACE1. These later observations were further confirmed by both activity assays and siRNA knockdown. NPD1 reduces Ab42 levels released from HNG cells over-expressing APPsw in a dose-dependent manner. Our examination of other bAPP fragments revealed after NPD1 addition, a reduction in the b-secretase products sAPPbsw and CTFb occurred, along with an increase in a-secretase products sAPPa and CTFa, while levels of bAPP expression remained unchanged in response to NPD1. Hence these abundance- and activity-based assays indicate a shift by NPD1 in bAPP processing from the amyloidogenic to non-amyloidogenic pathway. Previously sAPPa has been found to promote NPD1 biosynthesis from DHA, while in the present study NPD1 works to stimulate sAPPa secretion, creating positive feedback and neurotrophic reinforcement. Secreted sAPPa’s beneficial effects include enhanced learning, memory and neurotrophic properties. NPD1 further down-regulated the b-secretase BACE1 and activated ADAM10, a putative a-secretase. Our ADAM10 siRNA knockdown and BACE1 over-expression-activity experiments confirmed that ADAM10 and BACE1 are required in NPD1’s regulation of bAPP. NPD1 therefore appears to function favorably in both of these competing bAPP processing events. PPARc activation leads to anti-inflammatory, anti-amyloidogenic actions and anti-apoptotic bioactivity, as does NPD1. Some fatty acids are natural ligands for PPARc, which have a predilection for binding polyunsaturated fatty acids. Our hypothesis that NPD1 is a PPARc activator was confirmed by results from both human adipogenesis and cell-basedtransactivation assay. NPD1 may activate PPARc via direct binding or other interactive mechanisms. Analysis of bAPP-derived fragments revealed that PPARc does play a role in the NPD1-mediated suppression of Ab production. Over-expressing PPARc or incubation with a PPARc agonist led to reductions in Ab, sAPPb and CTFb similar to that with NPD1 treatment, while a PPARc antagonist abrogated these reductions. Activation of PPARc signaling is further confirmed by the observation that PPARc activity decreased BACE1 levels, and a PPARc antagonist overturned this decrease. Thus, the antiamyloidogenic bioactivity of NPD1 is associated with activation of the PPARc and the subsequent BACE1 down-regulation. The difference between the bioactivity of NPD1 concentrations for anti-apoptotic and anti-amyloidogenic activities may be due to the different cell models used and/or related mechanisms. Although Ab-lowering effects of PPARc have been reported, the molecular mechanism of this action remains unclear.
effects of NPD1 on PS1 levels, but a significant increase in ADAM10 coupled to a decrease in BACE1. These later observations were further confirmed by both activity assays and siRNA knockdown. NPD1 reduces Ab42 levels released from HNG cells over-expressing APPsw in a dose-dependent manner. Our examination of other bAPP fragments revealed after NPD1 addition, a reduction in the b-secretase products sAPPbsw and CTFb occurred, along with an increase in a-secretase products sAPPa and CTFa, while levels of bAPP expression remained unchanged in response to NPD1. Hence these abundance- and activity-based assays indicate a shift by NPD1 in bAPP processing from the amyloidogenic to non-amyloidogenic pathway. Previously sAPPa has been found to promote NPD1 biosynthesis from DHA, while in the present study NPD1 works to stimulate sAPPa secretion, creating positive feedback and neurotrophic reinforcement. Secreted sAPPa’s beneficial effects include enhanced learning, memory and neurotrophic properties. NPD1 further down-regulated the b-secretase BACE1 and activated ADAM10, a putative a-secretase. Our ADAM10 siRNA knockdown and BACE1 over-expression-activity experiments confirmed that ADAM10 and BACE1 are required in NPD1’s regulation of bAPP. NPD1 therefore appears to function favorably in both of these competing bAPP processing events. PPARc activation leads to anti-inflammatory, anti-amyloidogenic actions and anti-apoptotic bioactivity, as does NPD1. Some fatty acids are natural ligands for PPARc, which have a predilection for binding polyunsaturated fatty acids. Our hypothesis that NPD1 is a PPARc activator was confirmed by results from both human adipogenesis and cell-basedtransactivation assay. NPD1 may activate PPARc via direct binding or other interactive mechanisms. Analysis of bAPP-derived fragments revealed that PPARc does play a role in the NPD1-mediated suppression of Ab production. Over-expressing PPARc or incubation with a PPARc agonist led to reductions in Ab, sAPPb and CTFb similar to that with NPD1 treatment, while a PPARc antagonist abrogated these reductions. Activation of PPARc signaling is further confirmed by the observation that PPARc activity decreased BACE1 levels, and a PPARc antagonist overturned this decrease. Thus, the antiamyloidogenic bioactivity of NPD1 is associated with activation of the PPARc and the subsequent BACE1 down-regulation. The difference between the bioactivity of NPD1 concentrations for anti-apoptotic and anti-amyloidogenic activities may be due to the different cell models used and/or related mechanisms. Although Ab-lowering effects of PPARc have been reported, the molecular mechanism of this action remains unclear.
Importantly studies in Drosophila have provided additional insight into mechanisms in corals exposed to sudden environmental changes
In this respect, it is interesting to note that the heating rate explained more than 78% of the variation in Bcl2:Bax transcripts in A. millepora during our thermal stress experiments. This result suggests that the heating rate could be an important environmental driver of this molecular response in the host rather than duration and intensity of thermal anomalies. However, because the thermal treatments used in this study varied in both the average temperature and the heating rate, the effects observed on the level of expression of Bcl-2 family members are attributable to some combination of these 2 factors. Future studies should therefore include thermal treatments with tight control of temperature in order to precise the relative contributions of each factor on the molecular regulation of apoptosis in corals. This study provides further evidence that Tulathromycin B corals can regulate apoptosis in response to thermal stress. Bleaching is nevertheless an extremely dynamic event with many different triggers, responses and consequences to coral-dinoflagellate symbiosis. Understanding the cellular dynamics and the mechanisms driving the breakdown of this symbiosis will require many further studies but this is a critical step to predict future bleaching events and coral mortality under a changing climate. The formation of embryonic tissues is a key feature in generating diversity in animal development. After cell fate is established, cell-cell signaling and intracellular signal transduction pathways instruct cells to undergo cell shape changes. These cell shape changes are necessary for cell movement, a basic process that underlies embryonic development and is largely accomplished by regulation of the actin cytoskeleton. Actin dynamics is required for the migration of individual groups of cells, as in border cell migration in the Drosophila ovary, or large groups of cells, such as those involved in gastrulation in the developing fly embryo. One common feature of cell rearrangements via the actin cytoskeleton is the involvement of the Rho family of GTPases. Widely conserved across species and involved in seemingly diverse developmental processes including cell migration, phagocytosis, and myoblast fusion, the Rho GTPases are key signaling molecules that impinge upon actin cytoskeletal reorganization. Several classes of GTPase regulatory proteins have been identified, including the GTPase-activating proteins, guanine nucleotide exchange factors, and guanine nucleotide dissociation inhibitors. In particular, the GEFs regulate GTPase activity by exchanging the inactive, GDP-bound Rac to the active, GTP-bound state. It is thought that GEFs are a crucial intermediate that signal from upstream cell surface receptors to mediate GTPase activation. Some GEFs directly associate with membrane receptors, while others are associated via an intermediate complex. In flies, two neuronally expressed Rac GEFs have been identified that exemplify this in development of the central nervous system. Trio physically interacts with the Netrin receptor Frazzled to regulate chemoattraction, while Son of sevenless associates with the Roundabout receptor through the SH2-SH3 adaptor protein Dreadlocks to control axon repulsion. Vertebrate cell culture studies show this region is required for membrane localization. In flies, the DHR1 domain  is not essential for recruitment to the membrane, but is essential for myoblast fusion as deletion of the DHR1 domain fails to rescue mbc mutant embryos in functional rescue assays. Although the SH3-domain containing protein Crk is capable of binding the C-terminal proline-rich region of both DOCK180 and Mbc, it is not 3,4,5-Trimethoxyphenylacetic acid always essential in vivo. A direct interaction between vertebrate DOCK180 and CrkII is not required for apoptotic cell removal. Conventional GEFs bind nucleotide-free Rac via their DH domain, while the CDM proteins use the DHR2 region. Deletion or mutation of this domain results in a loss of Rac binding and activation. A DOCK-Rac protein complex is sufficient for Rac activation, but may be enhanced by DOCK180 bound to ELMO.
is not essential for recruitment to the membrane, but is essential for myoblast fusion as deletion of the DHR1 domain fails to rescue mbc mutant embryos in functional rescue assays. Although the SH3-domain containing protein Crk is capable of binding the C-terminal proline-rich region of both DOCK180 and Mbc, it is not 3,4,5-Trimethoxyphenylacetic acid always essential in vivo. A direct interaction between vertebrate DOCK180 and CrkII is not required for apoptotic cell removal. Conventional GEFs bind nucleotide-free Rac via their DH domain, while the CDM proteins use the DHR2 region. Deletion or mutation of this domain results in a loss of Rac binding and activation. A DOCK-Rac protein complex is sufficient for Rac activation, but may be enhanced by DOCK180 bound to ELMO.
Mutations in mbc and elmo result in border cell migration defects in the ovary and myoblast fusion defects in the embryo
Decreased Mbc and ELMO function exhibit abnormal ommatididal organization in the eye and thorax closure defects in the adult. In addition, loss-of-function studies have Ginsenoside-Ro demonstrated that the Rac genes are required redundantly in a variety of developmental processes, including border cell migration, myoblast fusion, and axon guidance in the developing nervous system. Last, genetic interactions exist between the atypical GEF Mbc-ELMO complex and their target GTPase Rac. A genetic screen in the eye uncovered an allele of mbc that suppresses the Rac1 overexpression phenotype. In support of this, removal of one copy of both Rac1 and Rac2 are capable of ameliorating the ”activated-Rac” phenotype exhibited by co-expression of both Mbc and ELMO in the eye. Although the work cited above provides convincing evidence that the DOCK180/Mbc-ELMO complex is essential in development, the mechanism by which at least five Rac-specific DOCK proteins bind to one or more ELMO proteins in vertebrates to modulate actin regulation in a tissue-specific manner is not clear. DOCK180, DOCK4, and DOCK5 are broadly expressed in many tissues, including the brain and nervous system. In contrast, DOCK2 is expressed specifically in hematopoietic cells, while DOCK3 expression is primarily restricted to the brain and spinal cord. In 3,4,5-Trimethoxyphenylacetic acid addition to their complex expression patterns, DOCK family members exhibit pleiotropic functions in development. DOCK180 has recently been shown to be required for Rac-mediated axon outgrowth in cortical neurons in response to netrin-1, neurite outgrowth as mediated by nerve growth factor, and axon pruning via ephrin-B3. Mouse knock-outs show DOCK180 is required in concert with DOCK5 in muscle fusion. DOCK3 colocalizes with N-cadherin and actin in neuronal differentiation. MOCA is  also linked to Alzheimer’s disease, where it accumulates in neurofibrillary tangles and modulates betaamyloid precursor processing. Consistent with this, mice lacking DOCK3 exhibit axonal degeneration. Finally, knockdown of DOCK4 results in reduced dendritic growth and branching in hippocampal neurons. Drosophila provides an excellent system to characterize this conserved pathway with a single ELMO ortholog. Using proteomics approaches for identifying new players in the ELMO-mediated pathway in the developing embryo, we have uncovered Spg, the Drosophila ortholog of human DOCK3/4, as an ELMO-interacting protein. In contrast to the well-established role of Mbc in myoblast fusion, Spg is not required with ELMO in somatic muscle development. However, the two Drosophila DOCK family members Mbc and Spg are required in the developing nerve cord. Moreover, Spg can be recruited to the membrane by Ncadherin in S2 cells, providing a mechanism for Spg localization that may function to mediate the development of axonal pathways. In the musculature, the only known GEF shown to be required for Rac activation is the Mbc-ELMO complex. However, in the developing nervous system, in addition to the unconventional DOCK-ELMO complexes, the conventional GEFs Trio and Sos are required. It is not clear how these multiple GEFs are regulated throughout CNS development. Possible mechanisms include the: regulation of GEF expression either in subsets of specific neurons or precise subcellular localization within the same neuron; unique physical associations between GEFs and receptors specific for distinct steps in axonal patterning; and regulation of GEF activity via post-translational modifications including phosphorylation or ubiquitination. While these ideas have not been examined in detail for all known GEFs, what is known is discussed below. First, it is possible mechanisms exist within the cell or tissue to compartmentalize GEF function as the spatial expression patterns of all GEFs in the developing ventral nerve cord seems to be fairly broad. Mbc is expressed at low or undetectable levels with reagents currently available, while Spg is expressed in all commissural and longitudinal axons, but not glial cells.
also linked to Alzheimer’s disease, where it accumulates in neurofibrillary tangles and modulates betaamyloid precursor processing. Consistent with this, mice lacking DOCK3 exhibit axonal degeneration. Finally, knockdown of DOCK4 results in reduced dendritic growth and branching in hippocampal neurons. Drosophila provides an excellent system to characterize this conserved pathway with a single ELMO ortholog. Using proteomics approaches for identifying new players in the ELMO-mediated pathway in the developing embryo, we have uncovered Spg, the Drosophila ortholog of human DOCK3/4, as an ELMO-interacting protein. In contrast to the well-established role of Mbc in myoblast fusion, Spg is not required with ELMO in somatic muscle development. However, the two Drosophila DOCK family members Mbc and Spg are required in the developing nerve cord. Moreover, Spg can be recruited to the membrane by Ncadherin in S2 cells, providing a mechanism for Spg localization that may function to mediate the development of axonal pathways. In the musculature, the only known GEF shown to be required for Rac activation is the Mbc-ELMO complex. However, in the developing nervous system, in addition to the unconventional DOCK-ELMO complexes, the conventional GEFs Trio and Sos are required. It is not clear how these multiple GEFs are regulated throughout CNS development. Possible mechanisms include the: regulation of GEF expression either in subsets of specific neurons or precise subcellular localization within the same neuron; unique physical associations between GEFs and receptors specific for distinct steps in axonal patterning; and regulation of GEF activity via post-translational modifications including phosphorylation or ubiquitination. While these ideas have not been examined in detail for all known GEFs, what is known is discussed below. First, it is possible mechanisms exist within the cell or tissue to compartmentalize GEF function as the spatial expression patterns of all GEFs in the developing ventral nerve cord seems to be fairly broad. Mbc is expressed at low or undetectable levels with reagents currently available, while Spg is expressed in all commissural and longitudinal axons, but not glial cells.