Furthermore, the employed HIF binding data in this study is for the HIF1a subunit only, whereas transcription factor cooperativity may well apply to other HIF subunits. In fact, several reports have implicated the ETS family of transcription factors in target selection by HIF2a. We observed very similar tendencies when transcriptional activation of reporter constructs was elicited by DMOG or hypoxia treatment, additionally suggesting that, at least in our experimental conditions, the contribution of these factors could occur mainly in basal conditions, as it is unlikely that hypoxia and DMOG treatment induce completely overlapping cellular responses. Several recent reports have suggested that chromatin accessibility determines HIF1 binding, although this mechanism may not fully explain HIFs binding and target selectivity. Our results indicate that an additional layer of specificity comes from proximal co-binding of other transcription factors and HIFs to open chromatin regions, thereby Benzethonium Chloride facilitating or restricting HIF-mediated transcription. Elucidation of the underlying molecular 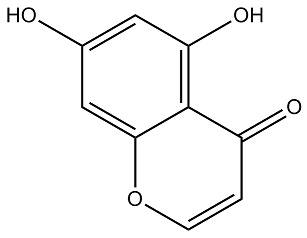 mechanisms falls outside the scope of our work, although it is tempting to speculate that transcription factors binding in proximity of HIFs may be involved in recruitment of co-activator or co-repressor proteins. Of note, a recent mammalian two-hybrid survey of protein-protein interactions for human and mouse TFs reported a physical association between HIF1A and AP-1 family member JUN, as well as the previously known interaction between CEBPB and p300. p300/CBP is a master co-activator of HIF-mediated transcription whose recruitment can also be mediated by CREB. In this regard, evidence from a synthetic transactivation screen on the EGLN1 promoter pointed to ETV4 as an additional p300-dependent coactivator of HIF-mediated transcription. Moreover, HIF1 is known to interact with Jab1/CSN5, a protein originally identified as a transcriptional coactivator for AP1. Future investigations on protein composition of HIF-bound enhancers should be pivotal in supporting this model. The associations between HIFs and AP1, CREB and CEBPs analyzed in our reporter results could be general across many HIF targets or be restricted to individual targets. To judge the generality of these results, we conducted a gene-set enrichment analysis of transcription factor targets in a sorted list of genes regulated by hypoxia. The results of this analysis showed a significant enrichment of CEBP targets among hypoxiainducible genes, suggesting that at least for this family of transcription factors, the functional association with HIFs could be relatively general. Of note, recent works have reported a direct protein-protein interaction between HIF1a and CEBPa, and have implicated CEBPa activity in regulation of the HIF target genes galectin-1 and PAI-1. Hypoxic induction of both galectin-1 and PAI-1 was found to be synergistically dependent on both HIF1a and CEBPa activity and their co-binding to the promoter region. Our results further suggest that this functional association may be general across a wider collection of HIF targets. In conclusion, the data presented herein demonstrates that integration of high-throughput chromatin immunoprecipitation and gene expression data is a successful approach to select highquality core HIF binding regions, and provides experimental proof of principle for the biological relevance of enriched transcription factor binding sites other than the HIF binding consensus in HIFmediated transcription. Specifically, our results suggest that Folinic acid calcium salt pentahydrate diverse stress-responsive transcription factors, in particular CEBPs, contribute to fine-tuning of the HIF-mediated transcriptional response. With the advent of the HIV epidemics, the disease has emerged as an important opportunistic infection in AIDS patients.
mechanisms falls outside the scope of our work, although it is tempting to speculate that transcription factors binding in proximity of HIFs may be involved in recruitment of co-activator or co-repressor proteins. Of note, a recent mammalian two-hybrid survey of protein-protein interactions for human and mouse TFs reported a physical association between HIF1A and AP-1 family member JUN, as well as the previously known interaction between CEBPB and p300. p300/CBP is a master co-activator of HIF-mediated transcription whose recruitment can also be mediated by CREB. In this regard, evidence from a synthetic transactivation screen on the EGLN1 promoter pointed to ETV4 as an additional p300-dependent coactivator of HIF-mediated transcription. Moreover, HIF1 is known to interact with Jab1/CSN5, a protein originally identified as a transcriptional coactivator for AP1. Future investigations on protein composition of HIF-bound enhancers should be pivotal in supporting this model. The associations between HIFs and AP1, CREB and CEBPs analyzed in our reporter results could be general across many HIF targets or be restricted to individual targets. To judge the generality of these results, we conducted a gene-set enrichment analysis of transcription factor targets in a sorted list of genes regulated by hypoxia. The results of this analysis showed a significant enrichment of CEBP targets among hypoxiainducible genes, suggesting that at least for this family of transcription factors, the functional association with HIFs could be relatively general. Of note, recent works have reported a direct protein-protein interaction between HIF1a and CEBPa, and have implicated CEBPa activity in regulation of the HIF target genes galectin-1 and PAI-1. Hypoxic induction of both galectin-1 and PAI-1 was found to be synergistically dependent on both HIF1a and CEBPa activity and their co-binding to the promoter region. Our results further suggest that this functional association may be general across a wider collection of HIF targets. In conclusion, the data presented herein demonstrates that integration of high-throughput chromatin immunoprecipitation and gene expression data is a successful approach to select highquality core HIF binding regions, and provides experimental proof of principle for the biological relevance of enriched transcription factor binding sites other than the HIF binding consensus in HIFmediated transcription. Specifically, our results suggest that Folinic acid calcium salt pentahydrate diverse stress-responsive transcription factors, in particular CEBPs, contribute to fine-tuning of the HIF-mediated transcriptional response. With the advent of the HIV epidemics, the disease has emerged as an important opportunistic infection in AIDS patients.
Monthly Archives: June 2019
The expression of fourteen genes in individual samples by real time qPCR
The high coefficient of determination obtained confirmed the high reliability of the microarray approach. This value was even shifted up when genes displaying low expression, and therefore less reliable Ct values in the correlation were excluded. Gonad samples were collected from Pacific oysters originating from 3 different sampling sites, at the four stages of the yearly reproductive cycle of oysters. This sampling method was undertaken in order to compensate for a possible bias in Atropine sulfate transcriptome analysis due to singularity of a single population within a precise environmental context. The results were mainly obtained from studying transcriptomic profiles from site 1 individuals, and show that the transcriptome of samples collected from different geographical locations are not significantly different regarding gametogenesis. Thus, we compared the transcriptomes of gonads from site 1 and site 2 using both microarray and real time qPCR analyses. The high correlation between geographical locations confirmed that the expression profiles observed are real features of gametogenesis in oysters without significant influence of the sampling site. Our analysis provided lists of genes Catharanthine sulfate expressed in male and female gonads, genes that increase in expression along the gametogenetic cycle, genes expressed in the flagella structure of spermatozoids, genes expressed in oocytes and genes expressed by female somatic cells. Most importantly, cross-referencing these lists of genes allowed us to identify potential markers of early sex differentiation in C. gigas oyster, a singular alternative hermaphrodite mollusk. We also provided new highly valuable information on genes specifically expressed by mature spermatozoids and mature oocytes. We  initially hoped to use PCA to discriminate between male and female stage 0 oysters and to identify genes involved in early sex differentiation. However, no difference was observed between the eight stage 0 gonads analyzed. Therefore, the future sexual development of these gonads could not be predicted from this PCA analysis. Principal component analysis revealed that differences between males and females increased overtime, from stage 1 gonads to stage 3 gonads, suggesting that sex differentiation takes place sometime before. Interestingly, some of the studied stage 0 gonads were found to express male specific or female specific genes, suggesting that sex differentiation already took place within these individuals although it was not possible to sex them using histology. We did not observe mitosis and cell proliferation within these individuals by histological methods. However, we performed cytology by observing a single transverse section of the gonad collected in the middle of the organ. Heterogeneity in the development of germ cells at different levels of the gonad has been observed and this may explain that some sex specific genes are found expressed when gene expression is measured on the whole gonad. Thus, the individuals presumed to be at stage 0 according to histological characterization, may biologically correspond to early stage 1 individuals. Few differences were observed between the transcriptomic profiles of undifferentiated stage 0 and sexed stage 1 individuals. Differences seem to fit with germ cell proliferation and the onset of meiosis in stage 1. The mitosis/ meiosis and sperm/oocyte decision may take place during the same time frame in the oyster gonad. In C. elegans, fbf and gld-3 genes control both the decision to leave the mitotic cell cycle and enter meiosis and to achieve the switch from spermatogenesis to oogenesis.
initially hoped to use PCA to discriminate between male and female stage 0 oysters and to identify genes involved in early sex differentiation. However, no difference was observed between the eight stage 0 gonads analyzed. Therefore, the future sexual development of these gonads could not be predicted from this PCA analysis. Principal component analysis revealed that differences between males and females increased overtime, from stage 1 gonads to stage 3 gonads, suggesting that sex differentiation takes place sometime before. Interestingly, some of the studied stage 0 gonads were found to express male specific or female specific genes, suggesting that sex differentiation already took place within these individuals although it was not possible to sex them using histology. We did not observe mitosis and cell proliferation within these individuals by histological methods. However, we performed cytology by observing a single transverse section of the gonad collected in the middle of the organ. Heterogeneity in the development of germ cells at different levels of the gonad has been observed and this may explain that some sex specific genes are found expressed when gene expression is measured on the whole gonad. Thus, the individuals presumed to be at stage 0 according to histological characterization, may biologically correspond to early stage 1 individuals. Few differences were observed between the transcriptomic profiles of undifferentiated stage 0 and sexed stage 1 individuals. Differences seem to fit with germ cell proliferation and the onset of meiosis in stage 1. The mitosis/ meiosis and sperm/oocyte decision may take place during the same time frame in the oyster gonad. In C. elegans, fbf and gld-3 genes control both the decision to leave the mitotic cell cycle and enter meiosis and to achieve the switch from spermatogenesis to oogenesis.
Understanding of the molecular mechanisms underlying the course of a reproductive cycle of oysters by describing their gonad transcriptome
Therefore, this study was designed to provide a better to establish lists of genes of interest specific to each reproductive 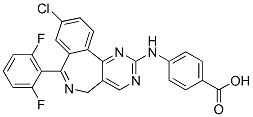 stage and sex. We employed a custom oligonucleotide microarray containing 31,918 ESTs Gomisin-D described and validated in Dheilly et al.. Our study identifies novel sex specific molecular markers and genes differentially expressed over the different stages of the gametogenesis cycle of males and females. Five hundred and eleven genes decreased in expression along the gametogenetic cycle. Numerous genes were previously identified as tissue-enriched in either the digestive gland, the mantle tissue, the visceral ganglion, hemocytes or the adductor muscle by Dheilly et al.. The method we employed did not exclude the possibility that genes that appear more expressed in early gonad developmental stages than in maturing gonads could be artifacts due to a dilution effect of genes expressed in somatic tissues, such as muscular fibers surrounding the gonadal tubules, when germ cells accumulate within the gonad area. In order to identify genes specifically expressed during early gametogenetic stages, we searched for genes significantly more expressed in immature gonads than in somatic tissues and mature gonads. Thus we compared expression data in stage 0 oyster gonads with expression data from somatic tissues previously described in Dheilly et al. in order to differentiate germline specific genes and genes somatically expressed in our gonad samples. Oyster gonad is a mixed tissue including storage tissue, smooth muscle fibers and circulating hemocytes. In order to characterize the expression of genes involved in gametogenesis in oocytes or female somatic tissue, we studied the transcriptome of oocytes collected by stripping 7 mature females and compared them to the transcriptome of the 10 stage 3 female gonad samples. Among the 2,482 genes differentially expressed in both male and female gametogenesis, 434 were significantly differentially expressed between female gonad tissue and stripped oocytes. Genes for which more transcripts were found in whole stage 3 gonads are predicted to be expressed by female somatic tissues. When more transcripts were found in stripped mature oocytes, the genes are predicted to be expressed by female germ cells. Predicted localization of gene expression is provided in file S3. Recent expression profiling studies using microarrays have provided great insight into the molecular mechanisms governing various complex physiological traits. Among those, the unprecedented amount of Butenafine hydrochloride information collected on gametogenesis, mitosis or meiosis of different eukaryotes such as yeast or nematode had a great impact on our understanding of sexual reproduction. Microarrays have also been employed successfully to better understand the cellular and molecular events of the development of reproductive tissues and of embryogenesis of cattle, mouse, rat and fish. Here, we proposed to unravel some molecular mechanisms involved in sex differentiation and gametogenesis of a peculiar alternative hermaphrodite invertebrate, the Pacific oyster Crassostrea gigas. We used an oligonucleotide microarray composed of 31,918 ESTs to characterize the transcriptome of oyster gonads at different developmental stages. The microarray employed in this study had previously been used to describe the transcriptome of various tissues of C. gigas and the results were validated by showing a significant correlation of gene expression obtained by real time qPCR and microarrays.
stage and sex. We employed a custom oligonucleotide microarray containing 31,918 ESTs Gomisin-D described and validated in Dheilly et al.. Our study identifies novel sex specific molecular markers and genes differentially expressed over the different stages of the gametogenesis cycle of males and females. Five hundred and eleven genes decreased in expression along the gametogenetic cycle. Numerous genes were previously identified as tissue-enriched in either the digestive gland, the mantle tissue, the visceral ganglion, hemocytes or the adductor muscle by Dheilly et al.. The method we employed did not exclude the possibility that genes that appear more expressed in early gonad developmental stages than in maturing gonads could be artifacts due to a dilution effect of genes expressed in somatic tissues, such as muscular fibers surrounding the gonadal tubules, when germ cells accumulate within the gonad area. In order to identify genes specifically expressed during early gametogenetic stages, we searched for genes significantly more expressed in immature gonads than in somatic tissues and mature gonads. Thus we compared expression data in stage 0 oyster gonads with expression data from somatic tissues previously described in Dheilly et al. in order to differentiate germline specific genes and genes somatically expressed in our gonad samples. Oyster gonad is a mixed tissue including storage tissue, smooth muscle fibers and circulating hemocytes. In order to characterize the expression of genes involved in gametogenesis in oocytes or female somatic tissue, we studied the transcriptome of oocytes collected by stripping 7 mature females and compared them to the transcriptome of the 10 stage 3 female gonad samples. Among the 2,482 genes differentially expressed in both male and female gametogenesis, 434 were significantly differentially expressed between female gonad tissue and stripped oocytes. Genes for which more transcripts were found in whole stage 3 gonads are predicted to be expressed by female somatic tissues. When more transcripts were found in stripped mature oocytes, the genes are predicted to be expressed by female germ cells. Predicted localization of gene expression is provided in file S3. Recent expression profiling studies using microarrays have provided great insight into the molecular mechanisms governing various complex physiological traits. Among those, the unprecedented amount of Butenafine hydrochloride information collected on gametogenesis, mitosis or meiosis of different eukaryotes such as yeast or nematode had a great impact on our understanding of sexual reproduction. Microarrays have also been employed successfully to better understand the cellular and molecular events of the development of reproductive tissues and of embryogenesis of cattle, mouse, rat and fish. Here, we proposed to unravel some molecular mechanisms involved in sex differentiation and gametogenesis of a peculiar alternative hermaphrodite invertebrate, the Pacific oyster Crassostrea gigas. We used an oligonucleotide microarray composed of 31,918 ESTs to characterize the transcriptome of oyster gonads at different developmental stages. The microarray employed in this study had previously been used to describe the transcriptome of various tissues of C. gigas and the results were validated by showing a significant correlation of gene expression obtained by real time qPCR and microarrays.
Genome duplications that gave rise to specialized spidroins occurred after the separation
The overall classification of spidroins shown here agrees with previous reports. The poorer repertoire of spidroins found in the Mygalomorphae clade suggests. However, a MaSp2like gene has been found in the Mygalomorphae spider Avicularia juruenses. This spidroin was formerly considered an orbicularian synapomorphy, and provided evidence that spidroin paralogization occurred prior to the divergence of mygalomorph and araneomorph spiders, estimated at 240 million years ago. We used the presence of MaSps in Mygalomorphae as evidence that gene duplication 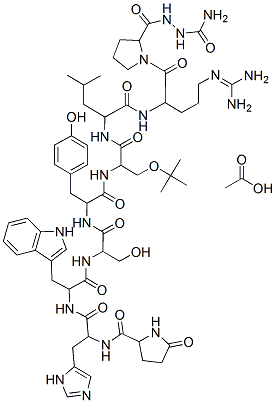 occurred before the anatomical specialization of glands. Regarding the evolution of spidroin gene families, our phylogeny suggests that pyriform spidroins are the sister group of the flagelliform spidroins and that aciniforms had a more recent genetic ancestor with the tubulliform spidroins. The poor bootstrap values observed the basis of the tree may indicate that most spidroin gene families originated in an almost simultaneous event of gene duplication that probably occurred after the separation between Mygalomorphae and the other spider clades. This event resulted in further subfunctionalization and neofunctionalization of spidroin gene families in some organisms and allowed the creation and usage of different sorts of silk, some more resistant and others more flexible. For example, MaSp1 presents a number of poly-A residues that give the silk more resistance. This spidroin is mainly used to build the first radial sustentation of webs. However, the MaSp2 and MiSp spidroins contain GPG and GGS motifs that give the web its elastic and sticky properties; this type of silk is used to fill in the radial parts of the web. The presence or absence of amino acid motifs in spider silk may arise from different motif duplications and reorganizations in the genome, allowing this sort of molecular neofunctionalization. Once they have occurred, these rearrangements may give a specific mechanical property to the web and allow the accomplishment of different tasks in a spider’s life. By translating the spidroin coding sequences from contigs and singlets into proteins, we detected the presence of insertions and deletions in their nucleotide sequences. We assumed that they had been added by problems in sequencing or base-calling procedures probably caused by the repetitive molecular nature of spidroins. However, it is likely that some spidroin genes will be revealed to have degenerated to pseudo-genes after genome duplications, as reported in other cases. In contrast, some of these duplicated genes have probably acquired new functions; a broad study of their duplication, neofunctionalization, subfunctionalization and decay associated with pleiotropy, fitness and mutational trade-offs would produce an interesting molecular evolutionary story. Here, a comprehensive transcriptomic analysis was conducted for the first time to evaluate the gene expression content of Folinic acid calcium salt pentahydrate spinning glands from two evolutionary distant spiders. The number of sequences Catharanthine sulfate evaluated in this study was more than 2.5 times larger than all the Araneae data previously deposited into the dbEST database. The sheer size of our dataset attests to the efficiency of NGS strategies for gene discovery projects, even for organisms lacking genomic information. We were surprised that the CAP3 software, with its simplistic method for sequence assembly developed over 10 years ago, produced a better performance in EST clustering than more recently developed and updated software.
occurred before the anatomical specialization of glands. Regarding the evolution of spidroin gene families, our phylogeny suggests that pyriform spidroins are the sister group of the flagelliform spidroins and that aciniforms had a more recent genetic ancestor with the tubulliform spidroins. The poor bootstrap values observed the basis of the tree may indicate that most spidroin gene families originated in an almost simultaneous event of gene duplication that probably occurred after the separation between Mygalomorphae and the other spider clades. This event resulted in further subfunctionalization and neofunctionalization of spidroin gene families in some organisms and allowed the creation and usage of different sorts of silk, some more resistant and others more flexible. For example, MaSp1 presents a number of poly-A residues that give the silk more resistance. This spidroin is mainly used to build the first radial sustentation of webs. However, the MaSp2 and MiSp spidroins contain GPG and GGS motifs that give the web its elastic and sticky properties; this type of silk is used to fill in the radial parts of the web. The presence or absence of amino acid motifs in spider silk may arise from different motif duplications and reorganizations in the genome, allowing this sort of molecular neofunctionalization. Once they have occurred, these rearrangements may give a specific mechanical property to the web and allow the accomplishment of different tasks in a spider’s life. By translating the spidroin coding sequences from contigs and singlets into proteins, we detected the presence of insertions and deletions in their nucleotide sequences. We assumed that they had been added by problems in sequencing or base-calling procedures probably caused by the repetitive molecular nature of spidroins. However, it is likely that some spidroin genes will be revealed to have degenerated to pseudo-genes after genome duplications, as reported in other cases. In contrast, some of these duplicated genes have probably acquired new functions; a broad study of their duplication, neofunctionalization, subfunctionalization and decay associated with pleiotropy, fitness and mutational trade-offs would produce an interesting molecular evolutionary story. Here, a comprehensive transcriptomic analysis was conducted for the first time to evaluate the gene expression content of Folinic acid calcium salt pentahydrate spinning glands from two evolutionary distant spiders. The number of sequences Catharanthine sulfate evaluated in this study was more than 2.5 times larger than all the Araneae data previously deposited into the dbEST database. The sheer size of our dataset attests to the efficiency of NGS strategies for gene discovery projects, even for organisms lacking genomic information. We were surprised that the CAP3 software, with its simplistic method for sequence assembly developed over 10 years ago, produced a better performance in EST clustering than more recently developed and updated software.
From a more applied point of view understanding the mechanisms regulating the seasonal reproduce
This result suggested that the cells require some other factors to maintain their immature properties. In our study, a number of different culture conditions were tested on SP cells for optimization. SP cells grew better and retained the SP characteristics on MEF feeder cells or in conditioned medium from MEF feeder cells, just like the case with mouse TS cells. Several different growth factors were also tested on SP cells. Because FGF2 gave the best result, FGF2 was chosen for a supplement with our SP medium, HSM. Although vCTB cells can be isolated from villi at any stage of pregnancy for primary culture, they quickly cease proliferating and differentiate within about 5 days. Previous studies reported that proliferation in 1st trimester villous explant could be increased at 4�C5 weeks by 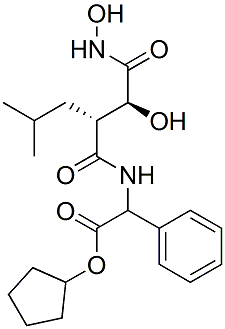 supplying EGF or IGF. FGF4 was also reported to inhibit differentiation of the 1st trimester explant and to prolong cell proliferation. Our SP cells from primary vCTB maintained the SP morphology in HSM containing FGF2 for at least 2 weeks after SP isolation. We did not check the later stage, but FGF2 is also a candidate mitogen for vCTB primary culture. We also discovered that IL7R and IL1R2 are novel markers of SP cells derived from both HTR-8/SVneo and primary vCTB. Most of the SP cells expressed IL7R and IL1R2, but in contrast, NSP cells failed to express IL7R and IL1R2. It is known that IL7R is expressed on Chloroquine Phosphate dendritic cells and monocytes, and activates multiple pathways that regulate lymphocyte survival, glucose uptake, proliferation and differentiation. IL7R signal in particular plays an essential role in T and B cell development and homeostasis. Although a previous study reported that IL7R was expressed in both vCTB and STB at an early stage of pregnancy its expression was rather weak. The function of IL7R in trophoblast differentiation remains unknown. The other marker, IL1R2, was reported to antagonize IL1 activity by acting as a decoy target for IL1 in polymorphonuclear cells, a specific type of leukocyte. IL1R2 expression has not been reported in placenta. Our in vitro data suggested that only a small proportion of vCTB cells expressed IL7R and IL1R2 and that they lost IL7R and IL1R2 expression as they differentiated into STB or EVT cells. Further investigation should reveal the function of IL7R and IL1R2 in the mechanism of human trophoblast differentiation. Isolation of human TS cells is necessary to investigate the early trophoblast cell lineages with self-renewing properties and the capability to differentiate into all trophoblast cell types of the mature placenta. The pathology of pregnancy-associated complication is believed to be based on abnormal trophoblast differentiation, defects in trophoblast invasion and spiral artery remodeling. To Tulathromycin B understand the pathology, in vitro model using human TS cells will provide tremendous benefits. Furthermore, human TS cells may lead us to a new approach for treating patients with placental dysfunction with TS cell transfer. Our study provides new insights into the characteristics of human TS/ progenitor cells. This study also reveals several key factors that are practical and available markers for TS/ progenitor cell isolation, and which might be essential for the maintenance of TS/ progenitor cells. These new insights should help us to understand human TS cell biology and develop novel therapeutic technologies for placental disorders. This offers a unique opportunity to investigate structure/function shifts during evolution and, by comparison with data from the two other bilaterian clades, to help define the basic assortment of genes required to manage reproduction.
supplying EGF or IGF. FGF4 was also reported to inhibit differentiation of the 1st trimester explant and to prolong cell proliferation. Our SP cells from primary vCTB maintained the SP morphology in HSM containing FGF2 for at least 2 weeks after SP isolation. We did not check the later stage, but FGF2 is also a candidate mitogen for vCTB primary culture. We also discovered that IL7R and IL1R2 are novel markers of SP cells derived from both HTR-8/SVneo and primary vCTB. Most of the SP cells expressed IL7R and IL1R2, but in contrast, NSP cells failed to express IL7R and IL1R2. It is known that IL7R is expressed on Chloroquine Phosphate dendritic cells and monocytes, and activates multiple pathways that regulate lymphocyte survival, glucose uptake, proliferation and differentiation. IL7R signal in particular plays an essential role in T and B cell development and homeostasis. Although a previous study reported that IL7R was expressed in both vCTB and STB at an early stage of pregnancy its expression was rather weak. The function of IL7R in trophoblast differentiation remains unknown. The other marker, IL1R2, was reported to antagonize IL1 activity by acting as a decoy target for IL1 in polymorphonuclear cells, a specific type of leukocyte. IL1R2 expression has not been reported in placenta. Our in vitro data suggested that only a small proportion of vCTB cells expressed IL7R and IL1R2 and that they lost IL7R and IL1R2 expression as they differentiated into STB or EVT cells. Further investigation should reveal the function of IL7R and IL1R2 in the mechanism of human trophoblast differentiation. Isolation of human TS cells is necessary to investigate the early trophoblast cell lineages with self-renewing properties and the capability to differentiate into all trophoblast cell types of the mature placenta. The pathology of pregnancy-associated complication is believed to be based on abnormal trophoblast differentiation, defects in trophoblast invasion and spiral artery remodeling. To Tulathromycin B understand the pathology, in vitro model using human TS cells will provide tremendous benefits. Furthermore, human TS cells may lead us to a new approach for treating patients with placental dysfunction with TS cell transfer. Our study provides new insights into the characteristics of human TS/ progenitor cells. This study also reveals several key factors that are practical and available markers for TS/ progenitor cell isolation, and which might be essential for the maintenance of TS/ progenitor cells. These new insights should help us to understand human TS cell biology and develop novel therapeutic technologies for placental disorders. This offers a unique opportunity to investigate structure/function shifts during evolution and, by comparison with data from the two other bilaterian clades, to help define the basic assortment of genes required to manage reproduction.